Experimental Guide of Sample Preparation for Flow Cytometry
Flow cytometry, also known as flow cytometry, is a very important bioanalytical technology and is commonly used in experimental scenarios such as cellular immune phenotype analysis, cell typing, and cell function assessment. Flow cytometry experiments mainly include four steps: preliminary experimental design, sample preparation, sample staining, and sample analysis. Sample preparation is one of the most important steps in flow cytometry experiments, which affects the accuracy of experimental results.
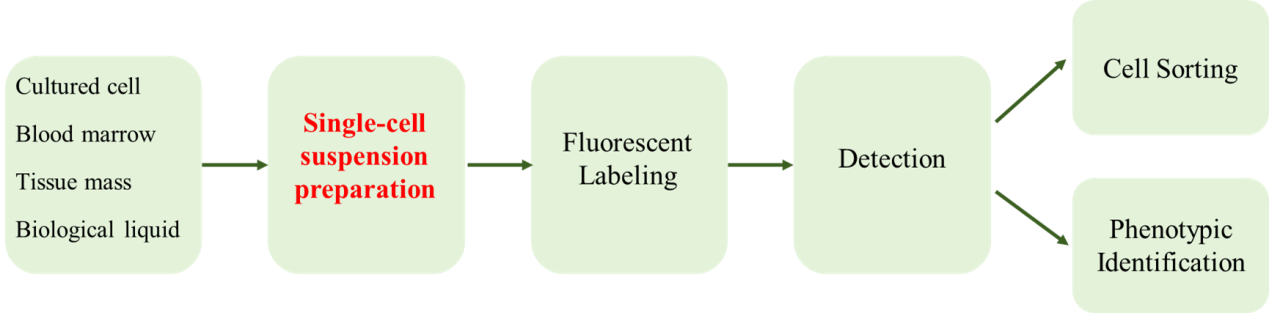
Figure 1 Main experimental process of flow cytometry
Single cell suspension is a prerequisite for cell analysis and detection by flow cytometry. During the preparation process, it not only requires the tissue to be dispersed into individual cells, but also maintains the inherent attributes and biological characteristics of the cells. Peripheral blood cells or cells growing in suspension are most suitable for flow cytometry analysis. There are many solutions available in the preparation process for other sample types. This article summarizes the experimental steps for preparing four types of flow cytometry samples into single cell samples for reference.
1. Peripheral whole blood
Peripheral whole blood anticoagulated with EDTA or sodium heparin is often used in flow cytometry experiments. Approach:
1. Use hemolysin to lyse red blood cells;
2. Use Ficoll or Percoll to isolate PBMCs from peripheral whole blood, and obtain the target cells from the corresponding density layer for cleaning and staining.
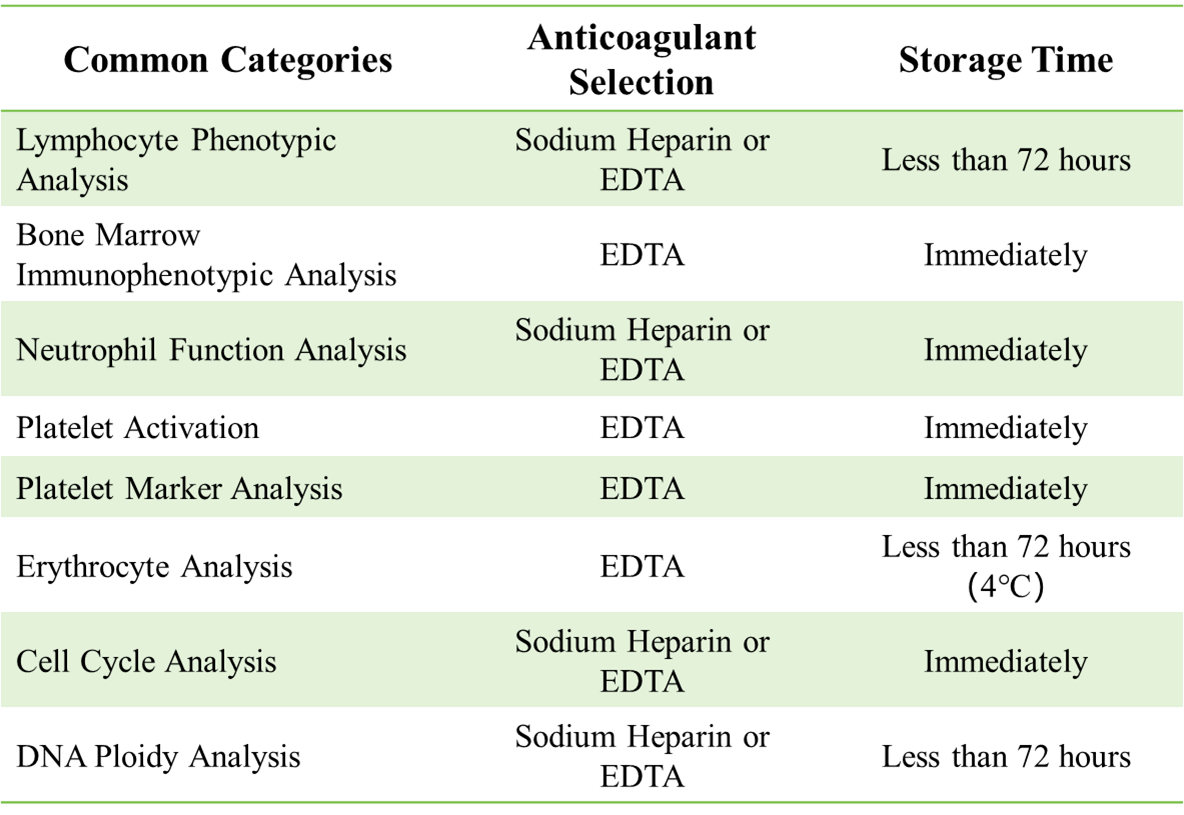
Table 1: Anticoagulant selection and storage time recommendations for blood samples

Table 2: Comparison of split red and lymphocyte separation solutions
1. Peripheral whole blood split red experimental protocol
1.1 Cracked Red Washing Plan
1) Add an appropriate amount of surface antibody to anticoagulated whole blood and incubate at room temperature for 15 minutes;
2) Add 1× red blood cell lysis solution (example R478427), incubate at room temperature in the dark for 10 minutes, centrifuge at 300-400 xg for 5 minutes, and remove the supernatant;
3) Add PBS/flow cytometry staining buffer (example T494526) and mix;
4) Centrifuge again according to the above conditions and disperse the pellet in PBS/flow cytometry staining buffer;
5) Test on the machine within 3 hours (if you cannot get on the machine immediately, resuspend the cells use 500 ul 4% paraformaldehyde (example P395744), store in the dark at 4°C, and analyze on the machine within 24 hours).
1.2 Cracked Red Washing Program
1) Add an appropriate amount of surface antibody to anticoagulated whole blood and incubate at room temperature for 15 minutes;
2) Add 1× red blood cell lysis solution and incubate at room temperature in the dark for 10 minutes;
3) Mix well and test on the machine.
2. Isolation of peripheral blood mononuclear cells
1) Dilute the blood sample 2-4 times with sterile diluent;
2) First add 15-20 mL of separation liquid into a 50mL conical bottom centrifuge tube, and then slowly add 20-30 mL of diluted whole blood or tissue cell suspension to the surface of the separation liquid;
3) Centrifuge at 400 xg for 15-30 minutes at room temperature to ensure that the rotor speed decreases smoothly;
4) Carefully take out the centrifuge tube from the centrifuge and slowly suck off the top layer to avoid contact with the mononuclear cell layer;
5) Slowly transfer the mononuclear cell layer to another 50 mL conical bottom centrifuge tube;
6) Add sterile washing solution and mix well, centrifuge at 300 xg for 10 minutes at room temperature, and carefully discard the supernatant;
7) To remove residual platelets, resuspend the cells in 30-50 mL sterile washing solution, centrifuge at 200 xg for 10-15 mins at room temperature, and discard the supernatant;
8) You can optionally repeat step 7 to remove most of the platelets;
9) Resuspend the cells in buffer or culture medium and perform subsequent operations such as detection and culture.
Note: The experimental steps for bone marrow samples are the same as those for peripheral whole blood.
3. Preparation of plasma enriched with platelets from peripheral blood
1) Take an appropriate amount of blood and centrifuge it at 300 xg for 10 minutes at 25°C;
2) Centrifuge the plasma layer obtained after centrifugation at 25°C and 1600 xg for 10 minutes;
3) Discard the supernatant and resuspend the pellet in Tyrode's buffer or EDTA-containing buffer for downstream experiments.
2. Culture cells
1. Collect cells in logarithmic growth phase.
2. Wash cells with PBS (example T494526).
3. Add trypsin (example T105532) or Accutase digestion solution to dissociate cells and filter through a mesh.
4. Centrifuge at 1500 rpm for 10 minutes and discard the supernatant.
5. Resuspend the cell pellet in PBS buffer (example T494526), collect the cell suspension, and perform cell counting and viability detection. Subsequent experiments can be carried out according to the procedures of fluorescent antibody staining.
3. Organization
1. Lymphoid tissue
1) Collect the spleen, thymus or lymph node into a cell culture dish containing buffer or culture medium, and grind it on the cell mesh with a syringe plunger;
2) Rinse the cell strainer with PBS (example T494526) to filter out cell clumps and debris;
3) Centrifuge at 300-400 xg for 5 minutes at 4˚C and discard the supernatant;
4) Resuspend the cells in an appropriate volume of buffer, perform cell counting and activity detection, and adjust the cell concentration to 107 cells/mL for downstream experiments.
2. Non-lymphoid tissue
1) Use surgical scissors or scalpel blades to cut the tissue into 2-4 mm pieces, and incubate with the appropriate type of digestive enzyme;
2) Pass the cell sieve and collect the cell suspension in a centrifuge tube;
3) Centrifuge at 300-400 xg at 4˚C for 5 minutes, discard the supernatant, and resuspend the cells in PBS (example T494526);
4) Repeat centrifugation, resuspend the cells in flow cytometry staining buffer, perform cell counting and activity detection, and adjust the final concentration to 10⁷ cells/mL.
Take the enzymatic treatment of solid tumor tissue as an example:
1. Preparation of solid tumor single cell suspension.
Enzymatic dissection of solid tumors utilizes trypsin to disrupt intercellular adhesions and type II collagenase to digest matrix components, and the addition of DNase I (example D128591) ensures that ploidy is determined only from intact cell populations. Enzymatic methods can be used alone or in combination with mechanical methods and are recommended for complete breakdown of squamous tumors.
1) Preparation of enzyme mixture: Add 2.5 mg/mL trypsin (example T105532), 0.5 mg/mL type II collagenase, and 20 µg/ml DNase I (example D128591) into a centrifuge tube, mix, and incubate at 37°C for 15 minutes;
2) Put fresh tumor tissue or mechanically decomposed tissue into the enzyme mixture and stir gently at 37°C for 1 hour;
3) Pass through an 80µm metal mesh sieve, add 2 ml of inactivated FBS to terminate the reaction, centrifuge at 250 xg for 10 minutes at room temperature, and discard the supernatant;
4) Resuspend the cells in culture medium, and perform cell counting and activity detection.
2. Ethanol fixation:
For solid tumor single cell suspensions that have been obtained, ethanol fixation can be used to extend the storage days without affecting nuclear integrity.
1) Vortex 2 mL of resuspended single cell suspension, while adding 6 mL of ice-cold 70% ethanol dropwise.
2) Cover and store at 4°C for 30 mins-5days.
3) The fixed cells can be used for some flow cytometry experiments.
4. Biological fluid samples
Such as alveolar lavage fluid, thoracic and ascites fluid, cerebrospinal fluid
1. Collect liquid samples, centrifuge at 300-400 xg for 5 minutes, discard the supernatant, and collect the cell pellet;
2. If red blood cells are visible to the naked eye in the cell pellet, add an appropriate amount of red blood cell lysis solution (example R478427), incubate in the dark for 2-10 minutes according to the red blood cell content of the sample, centrifuge at 300-400 xg for 5 minutes, discard the supernatant, and collect the cell pellet; resuspend the cells in PBS (example T494526) , centrifuge at 300-400 xg for 5 minutes and discard the supernatant (optional);
3.Resuspend the cells in PBS (example T494526) , centrifuge at 300-400 xg for 5 minutes, and discard the supernatant;
4. Resuspend the cells in an appropriate volume of PBS (example T494526) or flow cytometry staining buffer, perform cell counting and activity detection, and adjust the final concentration to 1x10 7 cells/ml for downstream experiments.
For more product details, please visit the Aladdin Scientific website.
