Enzyme Probes
Enzyme Probes
Enzymes have been used as detection probes for years and are still commonly used, even with the upsurge of fluorescent probes, because of their simplicity, sensitivity and broad-spectrum applications. Enzyme probes, including horseradish peroxidase (HRP) and alkaline phosphatase (AP), are used to detect target proteins through chromogenic, chemiluminescent or fluorescent outputs. The variability of these readouts demonstrates the versatility that enzymatic probes have in biological research methods, including immunohistochemistry (IHC), immunoblotting and enzyme-linked immunosorbent assays (ELISAs).
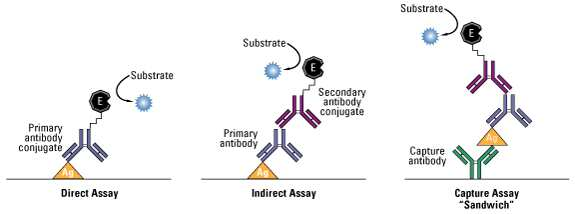
Common ELISA formats. In this assay, the antigen of interest is immobilized by direct adsorption to the assay plate or by first attaching a capture antibody to the plate surface. Detection of the antigen can then be performed using an enzyme-conjugated primary antibody (direct detection) or a matched set of unlabeled primary and conjugated secondary antibodies (indirect detection).
Introduction to enzyme probes
Enzymes as detection probe reporters
Enzymes are used to detect targets of interest by being conjugated to another protein or small molecule, such as a primary or secondary antibody, biotin, Avidin or some other protein or small molecule that binds to the target. The following illustration depicts Avidin-biotin complexes bound to antibodies as may occur in a typical immunoassay. A substrate of the specific enzyme is then introduced, which interacts with the enzyme to form a visible reaction product that can be a dark precipitate or the emission of light or fluorescence. This signal is then detected through microscopic or scanning methods.
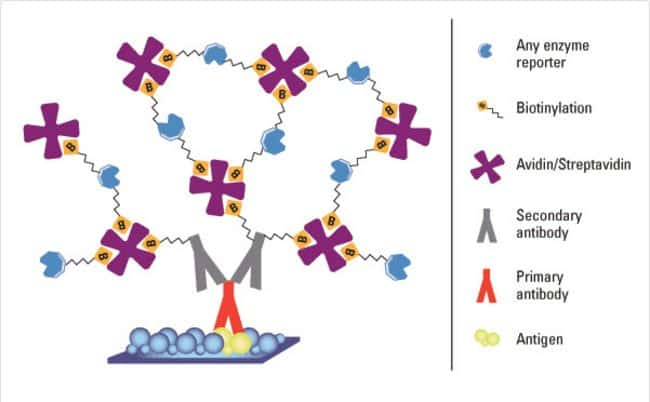
The benefits of using enzyme probes (also called reporters) to detect a target protein are three-fold:
High sensitivity – The signal output can be easily detected, and therefore low concentrations of target molecules can be identified. Additionally, methods of signal amplification are available that significantly increase the number of enzyme molecules to the site of the target molecule. Finally, enzyme reporters exhibit rapid turnover, which increases the amount of substrate that a single enzyme converts during a given unit of time.
Long shelf life – The enzymes are quite stable when stored properly, and while the enzyme substrate is light-sensitive, the enzyme itself is not sensitive to degradation by ambient light.
Output versatility – Substrates that yield either chromogenic, chemiluminescent or fluorescent output are available for the most common enzyme probes.
Although these benefits demonstrate the versatility and convenience of enzyme probes, there are limitations to their use that should be considered when choosing the appropriate type of detection probe:
Large size – Enzyme reporters are considerably larger than organic fluorescent compounds (e.g., FITC, TRITC, and AMCA) and therefore may interfere with the biological function of proteins to which they are conjugated.
Substrate requirement – Enzyme probes require the addition of a substrate for protein detection, and depending on the substrate used, this reaction can be sensitive to environmental conditions (e.g., light, temperature) and ambient light.
Endogenous interference – The enzymes used to detect target proteins in a sample are often expressed in the experimental system used (e.g., tissues, cells), which will also process the substrate and yield nonspecific background signal unless inhibited.
Common enzyme probe reporters and chromogenic substrates
Enzymes function as assay reporters by reacting with substrates to produce colored, fluorescent or chemiluminescent signals that can be measured. The table below lists several commonly used enzymes and substrates, which can be either soluble or precipitating. The illustration provides an example of the use of biotin in chromogenic detection.
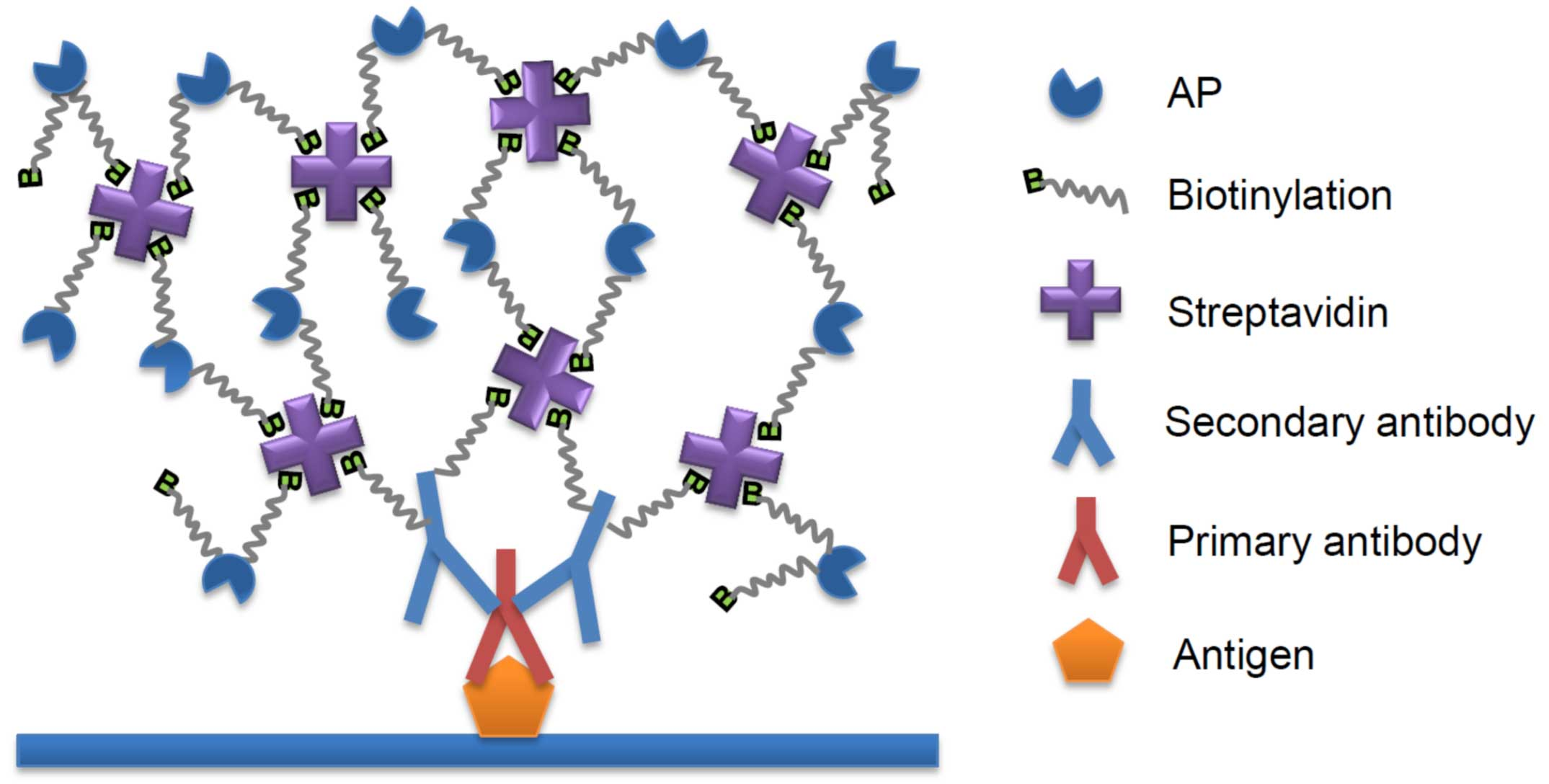
Biotin color detection. Tissue, cells, and antigens transferred to membranes or bound to porous cell culture plates first bind to the first antibody, which then binds to the biotin labeled second antibody; Mixing streptavidin and biotin labeled alkaline phosphatase in an appropriate proportion to form a network like SABC-AP complex; Then, the biotin labeled on the secondary antibody is combined with SABC-AP, and the AP substrate is added for color development to detect the corresponding signal. The key to signal amplification is the formation of a network like complex after mixing streptavidin and Biotin-AP, which is equivalent to the recognition of a biotin molecule to be detected through the SABC-AP complex, and ultimately the signal can be displayed by many AP molecules, achieving the effect of signal amplification.
Common enzyme probe reporters and chromogenic substrates
Enzymes function as assay reporters by reacting with substrates to produce colored, fluorescent or chemiluminescent signals that can be measured. Substrates can be either soluble or precipitating .
Enzyme label | Substrates |
Horseradish | 3,3'-diaminobenzidine (DAB) |
Alkaline | Combination of nitro blue tetrazolium chloride (NBT) |
Glucose oxidase | Nitro blue tetrazolium chloride (NBT) |
β-galactosidase | 5-bromo-4-chloro-3-indoyl-β-D-galactopyranoside (BCIG or X-Gal) |
Applications for enzyme probes
Enzyme probes are widely used in many different experimental applications because of their versatility, their ability to be conjugated to many different kinds of macromolecules and the availability and variability of substrates. Enzymes are most commonly used to detect proteins via direct and indirect antibody detection strategies, where the enzyme is conjugated to either the primary antibody that binds the target protein or the secondary antibody that targets the primary antibody, respectively. The enzyme can also be conjugated to biotin or Avidin for use in Avidin-biotin signal amplification systems.
Chromogenic detection using precipitating substrates is a common type of protein targeting used for immunohistochemistry (IHC), although in recent years the use of fluorescent probes has increased with technological advances and the greater availability of fluorescence detection instrumentation. Chemiluminescent substrates are the standard for protein detection by western blot analysis because of the high sensitivity and linear range of detection. Precipitating chromogenic substrates and, in recent years, fluorescent substrates are also available for this application. Chemiluminescent, fluorescent and soluble chromogenic ELISA substrates are commonly used, and each type of substrate exhibits characteristics specific for different experimental strategies.

Direct and indirect protein detection. The surface on which the target protein is bound and immobilized is either a membrane (western blot) or a microplate well (ELISA).
Enzyme reporters for probes
Horseradish peroxidase
Horseradish peroxidase (HRP) catalyzes the transfer of two electrons from a substrate to hydrogen peroxide to produce an oxidized substrate and water. For protein detection, HRP substrates (listed in the table below) are designed to generate a chromogenic, chemiluminescent or fluorescent signal upon oxidation. HRP has a molecular weight of 40,000, which is relatively small compared to other enzyme conjugates. This small size allows greater penetration into sample tissues and cells and reduces the likelihood of interfering with the conjugated protein function. HRP also has four lysines that are available for conjugation, which improves the efficiency of crosslinking to a protein of interest.
HRP has a high turnover rate and produces abundant reaction products in a short amount of time at physiological pH (7.6). HRP-IgG conjugates are superior to alkaline phosphatase and β-galactosidase conjugates due to their higher specific enzyme activity (more HRP/mole of antibody) and immunological reactivity (less steric hindrance because of the size of HRP).
One of the primary problems associated with the use of HRP is nonspecific staining that results from endogenous peroxide activity in certain tissues. Cryostat sections often contain a large amount of endogenous peroxidase activity, although commercial peroxidase inhibitors are available to reduce or eliminate endogenous peroxidase activity, HRP is the enzymatic label of choice for staining paraffin-embedded sections, a method that inhibits endogenous activity.
A second drawback of HRP is its sensitivity to degradation by microorganisms as well as the antibacterial agents used to fight them. Sodium azide is a potent inhibitor of HRP, but the enzyme can be stored in 0.01% thimerosal. HRP is also inhibited by cyanides, sulfides and azides. A final drawback of HRP is the mutagenicity or carcinogenicity of the reaction products of some horseradish peroxidase substrates. If any of these problems are a major concern, other enzymatic markers may be preferable.
Mechanism for detection with HisProbe-HRP probe. Thermo Scientific HisProbe-HRP is a nickel (Ni2+)-activated derivative of horseradish peroxidase (HRP) that enables direct, immobilized metal affinity chromatography (IMAC)-based detection of His-tagged proteins and other histidine-rich proteins in western blots and microplates.
Alkaline phosphatase
Alkaline phosphatases (AP) are a widely distributed family of enzymes that hydrolyze phosphates from nucleotides and proteins. Optimal enzymatic activity occurs at pH 9.0-9.6; these enzymes are activated by divalent cations and inhibited by cysteine, cyanides, arsenate, inorganic phosphate and divalent cation chelators, such as EDTA.
There are two forms of alkaline phosphatase in mammals; one form is distributed in a variety of tissues and the other is found only in the intestine. The two forms are affected differently by inhibitors and heat inactivators. The use of 1 mM levamisole in the substrate buffer will inhibit endogenous tissue alkaline phosphatase activity. Intestinal alkaline phosphatase activity may be inhibited by treating the sections prior to staining with 20% acetic acid at 4 °C for 15 seconds (or with 2.3% periodic acid for 5 minutes), followed by 0.02% potassium borohydride for 2 minutes.
Calf intestinal alkaline phosphatase conjugates are ideal in applications where high endogenous peroxidase levels contraindicate the use of HRP conjugates, such as with cryostat sections where peroxidase inhibitors are ineffective.
When used as a label, calf intestinal alkaline phosphatase (molecular weight 140,000) offers several distinct advantages over other enzymes. Because reaction rates remain linear, sensitivity can be improved by allowing the reaction to proceed for longer periods of time. The activity of calf intestinal alkaline phosphatase is not affected by exposure to antibacterial agents, such as sodium azide or thimerosal, so it may be stored for long periods of time in nonsterile environments. Because the endogenous activity of non-intestinal alkaline phosphatase can be inhibited by 1mM levamisole, antibodies labeled with this enzyme can be used as markers in many different tissues.
Glucose oxidase
Glucose oxidase is an enzyme isolated from Aspergillus niger that catalyzes the oxidation of β-D-glucose to produce hydrogen peroxide and gluconic acid. Glucose oxidase is a dimeric glycoprotein with a molecular weight of 160,000. Inhibitors of glucose oxidase include Ag+, Hg2+, Cu2+, and p-chloromercuribenzoate and phenylmercuric acetate.
Glucose oxidase is often the label of choice for samples with high endogenous peroxidase or phosphatase activity, as there is no endogenous glucose oxidase activity in mammalian tissues. It is important to choose a glucose oxidase with low catalase activity, though, because catalase destroys the hydrogen peroxide end product in the reaction.
β-galactosidase
β-galactosidase is an enzyme isolated from E. coli that is capable of hydrolyzing a variety of galactopyranoside derivatives, which produce both water-soluble and water-insoluble products. NaCl is an activator and Mg2+ is a cofactor of this enzyme, and the optimum pH for β-galactosidase is 7.0-7.5. For immunohistochemical staining, potential interference from endogenous enzyme can be overcome by embedding the sample with paraffin.
β-galactosidase is sensitive and demonstrates no endogenous activity in mammalian cells, and therefore it is useful in applications where endogenous enzyme activity is a persistent problem. β-galactosidase has been successfully coupled by a variety of crosslinkers to IgG fragments, whole immunoglobulins and insulin. One disadvantage of β-galactosidase is a lack of substrate variety.
Types of substrates
Chromogenic substrates
Chromogenic substrates have been widely used for many years and offer the simplest and most cost-effective method of detection. These substrates are divided into two groups based on the nature of the product of the enzyme-substrate reaction. When precipitating substrates in contact with the appropriate enzyme, they are converted to insoluble products that precipitate onto the sample or membrane. Because of the insoluble nature of these products, precipitating substrates are commonly used for IHC and western blotting. This representative data provides detailed information about a chromogenic western blot produced using an HRP-conjugated antibody and TMB solution.
Chromogenic western blot analysis Serial dilutions of HeLa cell lysate (7.5, 3.45, 1.88, 0.94, 0.47, 0.23, and 0.12 μg) were prepared and separated by electrophoresis. The proteins were transferred to nitrocellulose membranes and the membrane was blocked with 5 % skim milk in TBS + 0.05 % Thermo Scientific Tween 20. After blocking, the membrane was incubated with heat shock protein 86 polyclonal antibody at 0.5 μg/mL. The membrane was washed and then incubated with 0.2 μg/mL of HRP-conjugated goat anti-rabbit IgG and then washed again. The membrane was placed in 10 mL of 1-Step Ultra TMB-Blotting Solution and the color development was stopped at 5 minutes by rinsing the membrane with water.
Conversely, soluble substrates water-soluble colored products that dissolve into the test solution are commonly used in ELISA assays. Besides the difference in the nature of their products, these two types of chromogenic substrates differ in the detection instrumentation used. Precipitating substrates require no more specialized equipment than a light microscope to detect the presence, intensity and localization of the insoluble product, while a microplate reader is used to measure the absorbance and therefore the amount of soluble product in solution. Chromogenic blotting substrates are available in a wide variety of specifications and formats, and the appropriate substrate choice depends on the enzyme label, desired sensitivity and the form of the signal or method of detection needed.
For either type of chromogenic substrate, protein detection is similar to developing film; the sample is incubated in substrate until the color development is satisfactory, after which the reaction is halted and/or the amount of product measured. This approach allows the user to control the resolution of the data. The low sensitivity of chromogenic substrates, though, makes optimization of detecting low-abundance proteins difficult; although the reaction can be developed for several hours or even overnight, this also allows background signal to develop as well. Where chromogenic substrates fail in terms of sensitivity, they are ideal for applications where protein abundance is high. The signal is stable because the product of the substrate reaction is stable when stored properly, and hence chromogenic substrates do not typically have issues with false negative results that can occur with chemiluminescent substrates in Western blots (e.g., ghost bands).
Chromogenic substrates are typically used to detect abundant proteins, and reaction development can be monitored visually; this allows greater flexibility for optimization compared to chemiluminescent or fluorescent blotting systems. The performance of a particular substrate may vary dramatically when obtained from different suppliers, though, because performance can be affected by the concentration and purity of the substrate and by other additives and buffer components that are a part of the formulation.
Chemiluminescent substrates
Chemiluminescent substrates are popular because they offer several advantages over other detection methods, including a large linear response range that allows the detection and quantitation of a wide range of protein concentrations. Luminol is one of the most commonly used chemiluminescent reagents; luminol is oxidized by peroxide to form the excited-state 3-aminophthalate, which decays to a lower energy state by releasing photons of light at a wavelength of 425 nm.
Chemiluminescent substrates differ from other substrates in that the light detected is a transient product of the reaction that is only present while the enzyme-substrate reaction is occurring. This is in contrast to chromogenic substrates that produce a stable, colored product. In a chemiluminescent reaction, the substrate is the limiting reagent in the reaction; as it is exhausted, light production decreases and eventually ceases. A well-optimized procedure using the proper enzyme conjugate dilutions will produce a stable output of light for several hours, allowing consistent and sensitive detection of proteins.
Chemiluminescent substrates are commonly used for the short-term, rapid detection of protein detection by western blot analysis using x-ray film, phosphor imagers or CCD cameras.
Fluorescent substrates
While fluorophore-tagged antibodies and other molecules are more commonly used to detect target proteins, fluorescent substrates are also available for enzymatic detection. These substrates remain non-fluorescent or emit low fluorescence until metabolized by the enzyme probe, after which they emit intense fluorescence. These substrates offer greater sensitivity and the ability for rapid quantitation because of fluorescent microscopic imaging, fluorescent microplate readers and analytical software.
Enzyme labeling
Several methods have been developed and are used routinely to prepare antibody or other protein conjugates with enzymes such as HRP and AP. All kinds of enzyme-conjugated secondary antibodies are commercially available, but specialized primary antibodies and other protein probes may have to be labeled by the researcher.
Glutaraldehyde is one of the simplest crosslinking reagents used for protein conjugation. It reacts with amine groups to create crosslinks by one of several routes. Under reducing conditions, the aldehydes on both ends of glutaraldehyde couple with amines to form secondary amine linkages. The reagent is highly efficient at protein conjugation but has a tendency to form various high-molecular weight polymers, making results difficult to reproduce.
Periodate-activation followed by reductive amination is an excellent strategy for conjugation of HRP and other glycoproteins. Treatment of the glycosylated enzyme with periodate results in oxidation of sugar cis-diol groups (especially sialic acid, which is common in glycoprotein polysaccharides), resulting in formation of aldehyde groups. These aldehyde groups will then efficiently react (in the presence of the mild reductant cyanoborohydride) with primary amines of an added antibody or other molecule. The result is stable conjugation by permanent amide bonds.
Aldehydes (carbonyls) as crosslinking targets. The reaction of sodium periodate with sugar residues yields aldehydes for conjugation reactions. R and R' represent connecting sugar monomers of the polysaccharide. Red asterisks indicate sites of diol cleavage. Sialic acid is also called N-acetyl-D-neuraminic acid.
Recommended reading
[1] Sambrook J. and Russell D. W. (2001) Molecular cloning: A laboratory manual. Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory Press.
[2] CRC Handbook of Immunoblotting of Proteins: Volume 1 Technical Description. Eds Ole J. Bjerrum, Ph.D., M.D. and Niels H.H. Heegaard, M.D. CRC Press, Inc.: Boca Raton, FL, 1988.
[3] Bollag, D.M., et al. (1996). Protein Methods. Second Edition. Wiley-Liss, Inc., New York.
[4] Harlow, E. and Lane, D. (1988). Antibodies: A Laboratory Manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, New York.



