Preparation and precautions for single-cell suspensions of mouse tumor samples
Tumor sample preparation in mice
1. The tumor-bearing mice were killed by cervical dislocation method, soaked in 75% alcohol for 5 min, and then taken out and placed on a sterile operating table.
2. Holding forceps in the left hand and curved scissors in the right hand, cut a 1-cm-long slit along the edge of the tumor, so that the tumor can be clearly seen attached to the subcutis, and then gently cut open the connection along the edge of the tumor to peel off the tumor.
3. Place the stripped tumor into a 100 mm Petri dish and add 5~10 mL of 1640 basal medium at room temperature.
Preparation of single cell suspensions
A. Mixed enzyme digestion method
1. After all the tumors are stripped, transfer the tumors to 1.5 mL EP tube, hold the curved scissors to cut the tumors sufficiently, add 1640 basal medium while cutting, let it stand for a few seconds, use 1 mL pipette to suck out the upper layer of the smaller particles, continue to cut the tissues, and repeat the addition of 1640 basal medium until all the tissues meet the requirements in size.
2. Transfer the tumor tissue suspension to a 50 mL centrifuge tube, add 1640 basal medium, centrifuge at 250 g for 5 min, discard the supernatant, add 4.5 mL of 1640 basal medium, resuspend the cellular precipitate and transfer it to a petri dish.
3. Add 500 μL of 10×Triple Enzyme stock solution, gently blow until well mixed, and transfer to a 37℃ water bath shaker for digestion and incubation for 1~2 h. After digestion, incubate the cells with 1640 Enzyme stock solution.
4. At the end of digestion, dilute with 1640 basal medium or PBS, and then use a 200-mesh sieve to remove the residual tissue mass, and use 5-10 times the volume of 1640 basal medium or PBS buffer to wash once, and then filter until there is no tissue mass, to obtain the single-cell suspension.
5. Collect the cell suspension, centrifuge at 300 g for 5 min, and discard the supernatant.
6. Resuspend the cells with cell staining buffer and adjust the cell concentration to 1×107/mL.
B. Milling method
1. Prepare a 200-mesh disposable cell sieve mesh in advance, moisten it with 1640 basal medium or PBS and soak it for spare time (soak and place it on a 6 cm cell culture dish or 6-well plate).
2. Transfer the stripped tumor tissue to the 200-mesh cell sieve and cut it into small particles with sterile ophthalmic scissors.
3. Take the piston of a 2.5 mL syringe and grind the tissue with a soft tip in a circular motion until there are no visible tissue clumps on the sieve mesh, and rinse the sieve mesh 2~3 times with fresh 1640 medium or PBS.
4. Filter the obtained cell suspension through a 200-mesh cell sieve.
5. Collect the cell suspension, centrifuge at 300 g for 5 min, and discard the supernatant.
6. Resuspend the cells with cell staining buffer and adjust the cell concentration to 1×107/mL.
FSC/SSC plot of mouse tumor cells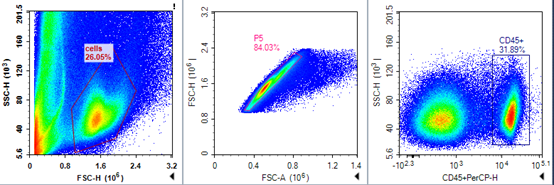
Mouse tumor cell CD45/SSC plot

Precautions:
1. Tumor volume should not exceed 1000 mm3 and tumor mass should be between 0.6 and 0.8 g. (If the tumor is large, the following reaction systems should be doubled.) (If the tumor is large, double the reaction system described below).
2. Enzymatic digestion should be performed with scissors so that the tumor can be freely aspirated with a 1 mL tip without obstruction.
3. To avoid over-digestion of the tissue during enzymatic digestion, take out the tissue every 20 min for observation, and end the digestion when there is no obvious tissue mass; the smaller the cell mass, the faster the digestion, and the tissue mass that can be blown with a 1 mL pipette can be digested in about 30-60 min; if the tissue is difficult to shear, the tissue can also be cut into sizes of about 1-2 mm3, and be blown with a 1 mL pipette every 30 min. Mixing once every 30 min, until it can be passed smoothly, about 1~2 hours can be fully digested. 4.
4. In order to minimize cell damage during grinding, it is necessary to soak in culture medium and avoid dry grinding.
For more information, visit our website: www.aladdinsci.com.
