Procainamide Labeling Kit-2-Picoline Borane-24T
Product description
Application: For labeling free glycans with procainamide (PROC).
Description: This kit contains reagents for conjugating dyes to the free reducing ends of glycans via reductive amination reactions.
Dye Properties: Massless Dye = 235.33.
Fluorescence, λex = 310 nm, λem = 370 nm.

Number of samples 12 individual assay samples per set of labeling reagents (24 samples total for kit)
Sample amounts per sample ranged from 25 pmol to 25 nmol glycans.
Appropriate samples can label any purified glycan with free reducing ends.
No detectable (< 2 mol %) loss of structural integrity sialic acid, fucose, sulfate or phosphate.
Label selectivity is essentially stoichiometric labeling.
Storage: Store at room temperature away from light. Keep away from heat, light sources and moisture. The reagents provided are stable for at least two years.
Shipping: This product can be shipped at ambient temperature.
Handling: Make sure any glass, plastic or solvent used is free of glycosidases and environmental carbons. Use powder-free gloves in all sample handling procedures and avoid contamination by environmental carbon compounds.
Safety: For Research Use Only. Not for human or drug use
Kit Contents
Each kit contains two labeling reaction sets. Each labeling reaction set consists of one vial of each of the following:
|
Additional reagents and equipment required
Pure water
Heat block, oven or similar dry heater (no water baths) set to 65°C
Centrifugal evaporators (eg Savant, Heto or similar)
Reaction vials (eg polypropylene microcentrifuge vials)
NOTE: Additional consumables are required if optional post-marking sample cleanup is performed (see sample cleanup section)
Timeline of labels
|
Labeling method
1 Purified glycans
It is best to remove non-carbon contaminants such as protein/peptide material, salts, and detergents from the sample prior to labeling glycans. This can be achieved using EB10 filter cartridges or protein-bound membranes. Purification of O-glycans can be achieved using CEX cartridges.
2 Transfer the sample to the reaction vial

The kit is designed to label up to 25 nmol of glycans per reaction. Using a single pure glycan, as low as 5 pmoles per reaction can be labeled and detected in subsequent HPLC and MS analysis.
Suitable reaction vials include small polypropylene microcentrifuge vials, PCR work vials, or 96 PCR plates for higher throughput sample analysis.
3.Dry samples and redissolve in 10 µL of water

If the sample volume exceeds 10 µL, dry the sample.
If the sample needs to be dried, it should be dried using a centrifugal evaporator.
If this is not possible, freeze drying (freeze drying) can be used with caution (especially to ensure that the sample is dried to a small dense mass at the bottom of the vial). Do not expose samples to high temperature (>28°C) or extreme pH conditions, as these conditions may lead to acid-catalyzed loss of sialic acid (high temperature, low pH) or glycan reducing end epimerization (at high temperature). under pH conditions).
After the samples were dried, 10 µL of water was added to redissolve the glycans.
4. Prepare the Dye Solution

Add 150 µl of kit component LT-ACETIC-DMSO-01 (30% acetic acid in DMSO) to a vial of procainamide dye and mix by pipetting until the dye is dissolved. Heat (30-60°C) is sometimes required to help dissolve the dye.
Transfer 150 µL of the dissolved dye solution to a bottle of reducing agent and mix by pipetting until the reducing agent dissolves. Sometimes heat (30-60°C) is needed to help
5 Add labeling reagent to the sample
Add 10 µl of labeling reagent to each glycan sample, cap the vial or PCR plate, mix thoroughly and centrifuge briefly (5-10 seconds) to ensure the labeling solution is at the bottom of the vial.
6 Incubation
Place the reaction vial in an oven or heat block set to 65°C and incubate for 1 hour. We recommend using an oven for the incubation step. The oven provides you with a full range of thermal reaction flasks.
The sample must be completely dissolved in the labeling solution for effective labeling. After 15 minutes of incubation to begin lysis, samples can be vortexed before continuing.
7 Cooling and Centrifugation
After the incubation period, remove the samples from the oven or heat block, allow them to cool to room temperature, and briefly centrifuge the reaction vial to ensure the sample is in the bottom of the vial and not in the lid.
Post-labeling sample cleanup
For most applications, we recommend that you perform post-label sample cleanup to remove non-glycan materials, such as excess dyes and other labeling reagents, prior to analysis by HPLC, MS, or LC-MS.
The benefits of post-label cleaning
Removes free dyes and chemicals that can interfere with HPLC and LC-MS sample analysis.
Extend the life of HPLC columns.
Smaller glycans such as O-link near the start of the HPLC gradient will be detected without interference from free dyes.
A full range of HPLC color columns can be used.
Sample capacity issues with HPLC color columns are unlikely to occur.
Analysis of procainamide-labeled glycans
Procainamide-labeled glycans can be studied by a number of different analytical methods, including (U)HPLC, ESI mass spectrometry, and LC-ESI-MS.
Example UHPLC conditions – Conditions vary by instrument. Check if these conditions are suitable for your device.
Samples are prepared in a solvent equivalent to the starting gradient, such as 76% nitrile.
Color column: Waters BEH Glycan color column 15 cm x 2.1 mm.
Column temperature: 40°C
Fluorescence detector settings: Excitation wavelength: 310 nm, Emission wavelength: 370 nm.
Solvent A: 50 mM ammonium formate buffer pH 4.4
Solvent B: Nitrile
Long UHPLC gradients for samples with unknown glycan profiles and glycans that may be large and/or highly sialylated.
This UHPLC gradient has been shown to separate N-glycans released from human IgG and erythropoietin, followed by procainamide labeling and cleanup. Once the range of glycans present in the sample is determined, the UHPLC gradient can be optimized and significantly shortened (to 15 minutes on some UHPLC instruments).
LC-MS analysis of procainamide-labeled N-glycans allows the user to obtain both fluorescence and ESI-MS spectra of the separated N-glycans, which can be combined to provide a larger overall presence A picture of the N-glycan structure. MS data can be used to assign m/z masses to each peak in the fluorochromatic spectrum, providing possible structural identities for each peak in the form of monosaccharide compositions. Through ESI-MS analysis, a single peak in the fluorescence spectrum can be determined to have multiple glycan structures.
Repeatability (precision) of procainamide labeling and purification procedure of N-glycans released from human IgG glycans showed low RSD values (RSD <5% for peaks with relative % area >0.7 %) .
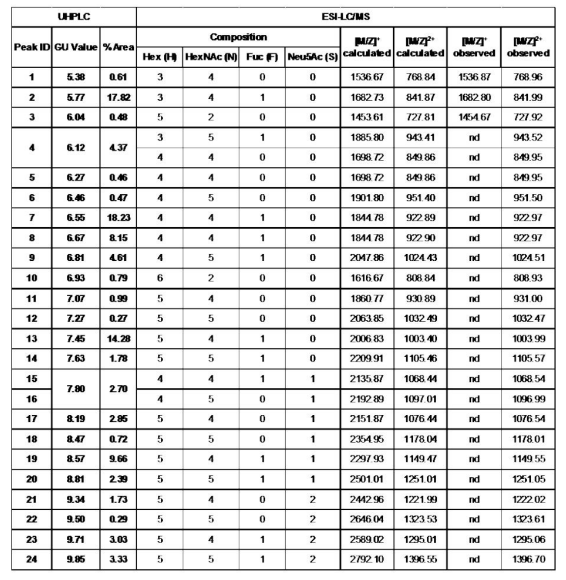
Structural annotations for each peak in this color spectrum of purified procainamide-labeled human IgG glycans
Purified procainamide-labeled erythropoietin glycans
Example ESI-MS spectrum.
Procainamide-labeled human IgG glycans. Approximately 10 pmol of glycans were injected into the LC-MS system. One sample, all data added. Mass spectra were run on a Bruker AmaZon Speed ETD instrument with positive ion settings.
Example ESI-MS spectrum of a procainamide-labeled IgG glycan peak.
The peaks are a mixture of Hex5HexNAc2 glycans: 727.93 [M+H]2+ (MAN5) and Hex3HexNAc4dHex1 glycans: 943.52 [M+H]2+ and 629.35 [M+H]3+ (FA2B). Inset shows base peak color spectrum (top curve) and fluorescence
Color spectrum (bottom trace). Arrows indicate the source of the MS spectrum.
Structural assignment of major procainamide-labeled N-glycans in human IgG.
Analysis of procainamide-labeled N-glycans by LC-MS allows users to assign tentative glycan structures to peaks in their samples.
Relative % peak area, GU value data and m/z value for each peak. Analysis of procainamide-labeled N-glycans by LC-MS allows users to specify tentative glycan structures
Mark repeatability (precision). Procainamide-labeled human IgG glycans after purification. Twenty independent glycan release and labeling samples were compared. normalized color spectrum
Relative % peak area comparison of samples in . Reproducibility (precision) of procainamide labeling and purification procedures for N-glycans released in humans. Low RSD values for IgG glycans (RSD <5% for peaks with relative % area >0.7 %)
Reductive amination reaction
1. Schiff base formation
This requires glycans with free reducing ends that balance between closed (cyclic) and open (acyclic) forms. The primary amino group of the dye conducts a nucleophilic attack on the carbonyl carbon of the acyclic reducing end residue, forming a partially stabilized Schiff
2. Reduce Schiff base
Schiff base imine groups are chemically reduced to produce stable labeled glycans
Glycans are labeled with procainamide (PROC) by reductive amination.
References
1. Kozak RP, Tortosa CB, Fernandez DL, Spencer DI. Comparison of procainamide and 2-aminobenzamide labeling for the analysis and identification of glycans by fluorescence detection by liquid chromatography coupled to electrospray ionization mass spectrometry. Anal biochemistry. 2015;486:38-40. doi: assay to analyze and identify glycans. Anal biochemistry. 2015;486:38-40. doi: 10.1016/j.ab.2015.06.006.
2. Klapoetke S, Zhang J, Becht S, Gu X, Ding X. Evaluation of a new method for the analysis and identification of procainamide-tagged N-linked glycans by HPLC with fluorescence and mass spectrometry detection. J Pharm Biomedical Anal. 2010;53(3):315-24. doi: 10.1016/j.jpba.2010.03.045.
3. . Pabst M, Kolarich D, Pöltl G, Dalik T, Lubec G, Hofinger A, Altmann F. Comparison of fluorescent labeling of oligosaccharides and introduction of new post-labeling purification methods. Anal biochemistry. 2009;384(2):263-73. doi: 10.1016/j.ab.2008.09.041.
4. Liu R, Giddens J, McClung CM, Magnelli PE, Wang LX, Guthrie EP. Evaluation of glycoengineered monoclonal antibodies by LC-MS analysis combined with multiple enzymatic digestions. Monoclonal antibodies. 2016;8(2):340-6. doi: 10.1080/19420862.2015.111336.