Product content
| F665904 | Component | 500 U | 2500 U | Storage | | F665904A | FastStar DNA Polymerase, 5 U/μL | 100 μL | 5×100 μL | -20℃. Avoid freeze/thaw cycle. | | F665904B | 10×PCR Buffer | 1.8 mL | 5×1.8 mL | -20℃. Avoid freeze/thaw cycle. |
Notes: 10×PCR Buffer contains 15mM Mg2+. |
Product Introduction FastStar DNA Polymerase is a mixture of anti-Taq enzyme monoclonal antibody and CW0680 Taq DNA Polymerase for Hot Start PCR.When using Taq enzyme antibody for PCR amplification, Taq enzyme antibody binds to Taq enzyme to inhibit the DNA polymerase activity prior to high temperature denaturation, and effectively inhibits non-specific annealing of primers and non-specific amplification caused by primer dimerization at low temperatures. It can effectively inhibit the non-specific annealing of primers and non-specific amplification caused by primer dimer under low temperature conditions.Taqase antibody denatures the DNA in the initial DNA denaturation step of the PCR reaction, and DNA polymerase activity is restored to achieve the effect of hot start. No special inactivation of Taq enzyme antibody is required for the use of this product, and it can be used under conventional PCR reaction conditions. FastStar DNA Polymerase with 5′→3′ DNA polymerase activity and 5′→3′ exonuclease activity, no 3′ →With 5′ exonuclease activity and an enzyme extension rate of 2kb/min, fragments up to 5kb in length can be amplified. The amplified PCR product has an "A" base at the 3′ end, so it can be directly used for T/A cloning. This product is characterized by fast extension speed and high amplification efficiency, and is mainly suitable for PCR amplification of DNA fragments, DNA sequence determination and other experiments. Active Definition Using activated salmon sperm DNA as template/primer, the amount of enzyme required to dope 10 nmol of deoxyribonucleotide into acid-insoluble material was defined as 1 activity unit (U) at 74°C for 30 min. quality control After several column purifications, its purity was greater than 99% by SDS-PAGE; no exogenous nuclease activity was detected; PCR method detects no host residual DNA; efficiently amplifies single-copy genes in the human genome. Usage The following example shows the PCR reaction system and reaction conditions for amplifying a 1kb fragment using human genomic DNA as a template, which should be improved and optimized according to the template, primer structure and size of the target fragment in actual operation. | Reagent | 50 μl Reaction system | final concentration | | 10× PCR Buffer | 5 μl | 1× | | dNTP Mix,10 mM each | 1 μl | 200 μM each | | Forward Primer,10 μM | 2 μl | 0.4 μM | | Reverse Primer,10 μM | 2 μl | 0.4 μM | | Template DNA | <0.5 μg | <0.5 μg/50 μl | | FastStar DNA Polymerase,5U/μl | 0.25-0.5 μl | 1.25-2.5 U/50 μl | | ddH₂O | up to 50 μl |
|
|
Note: The reaction solution can be prepared at room temperature; the reagents must be placed on ice. 2.PCR reaction conditions 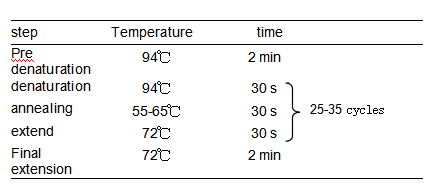
Attention: 1) In general, the annealing temperature in the experiment is 5°C lower than the melting temperature Tm of the amplification primer, and when the ideal amplification efficiency cannot be obtained, the annealing temperature is appropriately lowered. 2) The extension time should be set according to the size of the amplified fragment. 3) The number of cycles can be set according to the downstream application of the amplification product . If the number of cycles is too low, the amplification is insufficient; if the number of cycles is too high, the chance of mismatch will increase and the non-specific background will be serious. Therefore, the number of cycles should be minimized under the premise of ensuring the product yield. | 

