GAPDH catalyzes the conversion of glyceraldehyde-3-phosphate to 1, 3-bisphosphoglycerate (1,3-BPG), creating a high-energy phosphate bond while
simultaneously facilitating the formation of NADH. This is a key step in glycolysis. 1,3-BPG goes on to result in ATP production while the NADH powers the
electron transport chain, synthesizing more ATP. GAPDH is an essential constitutive enzyme whose activity is regulated by several factors, including cellular
energy demand, redox state and post-translational modifications. Dysregulation of GAPDH is associated with cancer and neurodegenerative disorders.
AkrivisBio’s GAPDH Assay is a simple, sensitive means of measuring GAPDH activity in a variety of samples with a sensitivity into the low µU/ml. Detection principle
- GAPDH converts glyceraldehyde-3-phosphate to 1, 3-bisphosphoglycerate with the conversion of NAD to NADH.
- NADH is used to reduce WST-8 from its nearly colorless tetrazolium form to a highly colored formazan. Storage and precautions
Storage and Handling Considerations:
Store unopened assay at -20°C.
Assay Buffer: Warm to room temperature (RT) before use. Store at 4°C.
Glyceraldehyde-3-phosphate: Ready to use as supplied. Aliquot into convenient portions and store at -20°C. Keep on ice while in use.
WST-8/NAD Mix: Reconstitute with 220 µl of DI H2O. Add and let it sit undisturbed for a few minutes, vortex briefly. Store at 4°C.
NADH Standard: Reconstitute with 400 μl of DI H2O giving a 0.5 mM solution. Aliquot into 35-40 µl portions and store at -20°C. Keep on ice while in use.
GAPDH Positive Control: Reconstitute with 100 μl of DI H2O. Aliquot and store at -70°C. Keep on ice while in use.
Test method
Assay Protocol:
1. Prewarm the plate reader to 37°C.
2. Standard Curve: Add 0 – 5 – 10 – 15 – 20 - 25 μl of the NADH Standard into a series of wells in a 96-well plate giving 0 – 2.5 – 5 – 7.5 – 10– 12.5 nmol of NADH. Adjust all well volumes to 50 μl with Assay Buffer.
3. Sample Preparation: Homogenize cells (1 x 106) or tissue (10 mg) with 100 µl Assay Buffer, on ice. Centrifuge at 16,000 x g, at 4°C for 5 minutes. Transfer all of the clear supernatant to a fresh tube. Add up to 50 μl sample and adjust sample well volumes to 50 µl with Assay Buffer.
4. Positive Control: Dilute 10 µl of the Positive Control by adding it to 90 μl of Assay Buffer. Transfer 2 – 5 μl to wells in the 96-well plate and adjust the well volume. to 50 µl with Assay Buffer. Place the plate in the plate reader to warm up while Rection Mix is prepared.
Note:Crude preps occasionally containotherenzymeswhichgenerateNADH. Ifthis is suspected, run samples in pairs with one used as a background control.
5. Reaction Mix: Each Standard, Positive Control, Sample and Background Control well will require 50 µl of Reaction Mix. Prepare sufficient material for the total number of wells to be run, containing:
| / |
Reaction Mix |
Background Control Mix |
| Assay Buffer |
46 µl |
48 µl |
| Glyceraldehyde-3-phosphate |
2 µl |
/ |
| WST-8/NAD Mix |
2 µl |
2 µl | |
add 50 µl of the Reaction Mix to each Standard, Positive Control and Sample well. Add Background Control Mix to any Background Control wells used.
6. Measurement: Immediately start measuring absorbance at 450 nm in kinetic mode for the wells being run. Acquire data for at least 30 minutes.
Typical Results 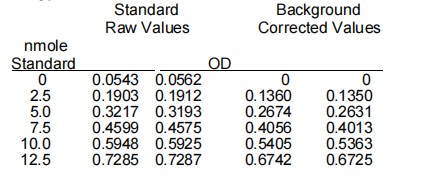
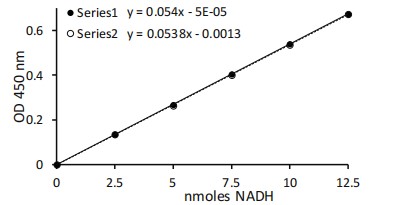
Calculation
Subtract the 0 Standard value from all Standard readings. Plot the NADH Standard Curve. Determine the slope of the standard
curve. Subtract any background control value from its paired sample reading. If there are no background controls, subtract the zero standard
from all sample values. Determine a linear range for the enzyme activity. Determine the slope for the linear portion in OD/minute. Divide the
slope of the activity by the slope of the standard curve to convert from OD/minute to nmoles/minute (mU). To convert from mU in the well to
activity in the original samples:
A. Divide the mU in the well by the volume of sample added to the well = mU per µl of sample
B. Multiply the mU per µl of sample by the total volume of supernatant recovered in step 3 above = total mU per sample
C. Divide total mU per sample by the mg tissue or # of cells of original sample = mU per mg tissue (or per # of cells, etc.)
| 


